PROMISE: Personalized Rehabilitation in Oncology specific Motor deficits using Intelligent Sensing and Extended reality
Context
Chemotherapy-induced neuropathies (CIPN) have gained clinical significance due to the prevalence of malignant disease and the use of new chemotherapeutic drugs; their prevalence is also reported to be 30%–40%, with high variance depending on the drug(s) used and treatment scheme. With over 600.000 new cases in 2018 and almost 2 million prevalent cases on a 5-year prediction in Germany, cancer is still in the foreground of chronic diseases that require innovative and impactful solutions. CIPN is one of the most frequent adverse effects of many commonly used chemotherapeutic agents and has a strong impact on patients’ quality of life. CIPN is a severe problem in oncology leading to dose reduction, treatment delay, or discontinuation. It causes long-lasting disturbances of daily functioning and quality of life in a considerable proportion of patients, due to its sensory and motor symptoms.
Research partners
Goal
PROMISE is a technological intervention for polyneuropathies that provides personalized quantification and adaptive compensation of sensorimotor deficits. The initial instantiation tackles CIPN and its rehabilitation in cancer survivors after chemotherapy. It proves the potential that motion capturing technologies, wearables, and machine learning algorithms have in combination in a digital intervention aiming at personalized physical rehabilitation.
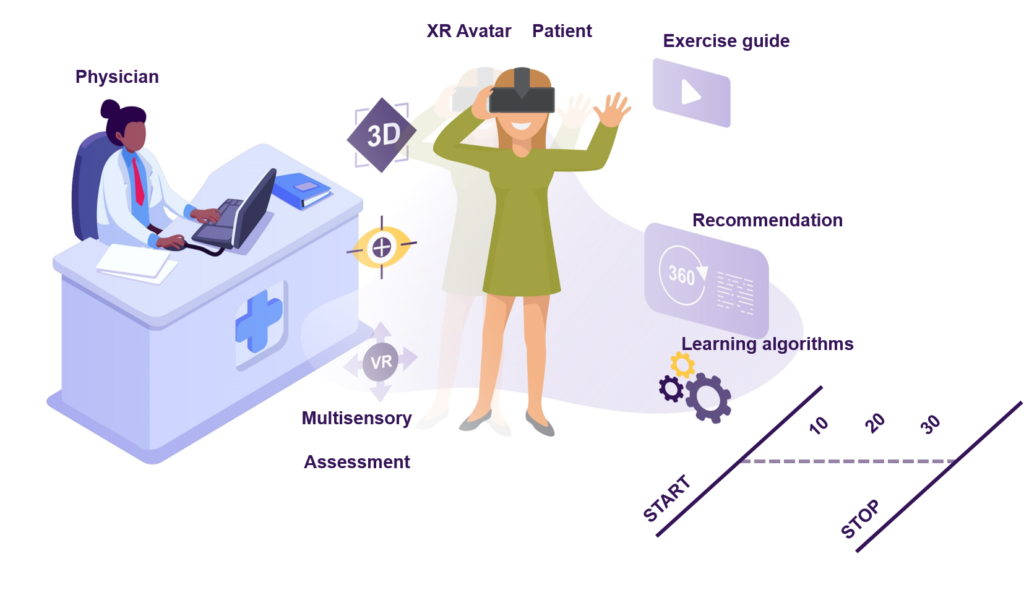
PROMISE explores the potential that digital interventions have for sensorimotor CIPN assessment and rehabilitation in cancer patients. Our primary goal is to demonstrate the benefits and impact that Extended Reality (XR) avatars and AI algorithms have in combination in a digital intervention aiming at 1) assessing the complete kinematics of deficits through learning underlying patient sensorimotor parameters, and 2) parametrize a multimodal XR stimulation to drive personalized deficit compensation in rehabilitation 3) quantitatively track the patient’s progress and automatically generate personalized recommendations 4) quantitatively evaluate and benchmark the effectiveness of the administered therapies.
Overview
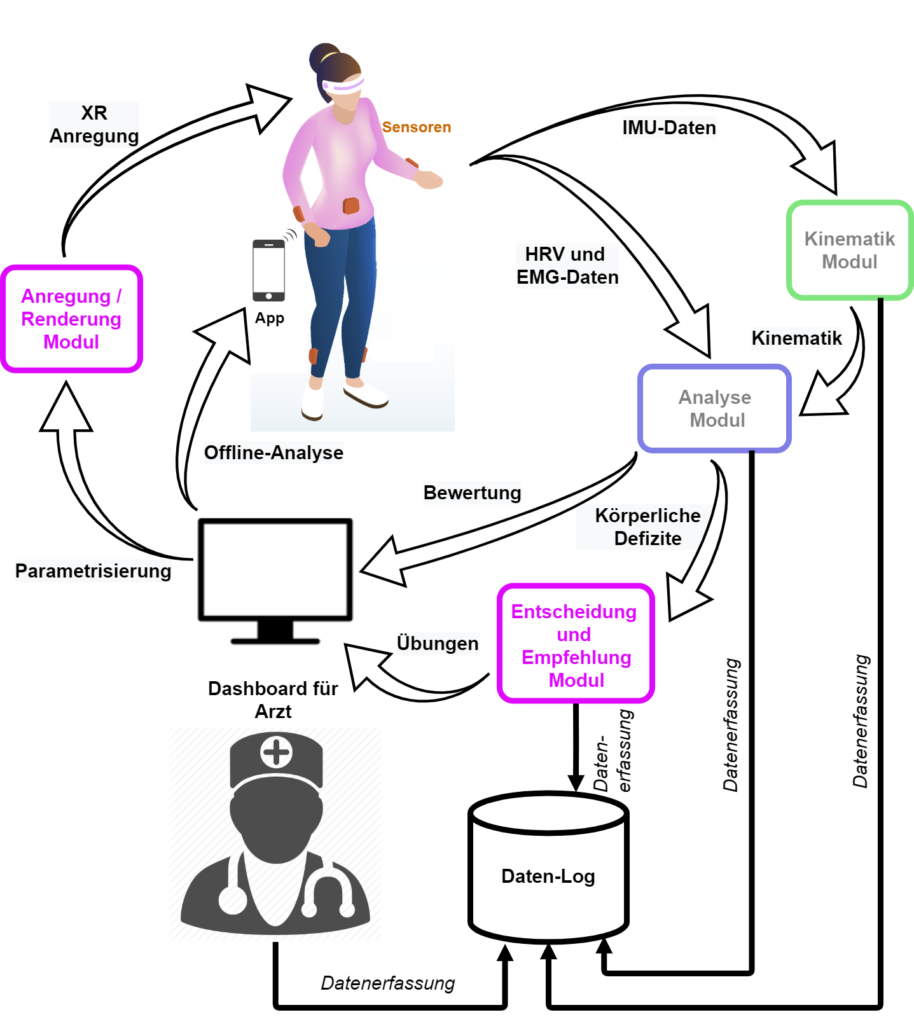
The generic system architecture is depicted in the following diagram. The rich sensory data is responsible to describe a complete assessment. Through machine learning algorithms each of the modules provides a powerful interface from the patient to the physician.
PROMISE will address the sensorimotor symptoms of CIPN. It will use commodity digital technologies, such as Extended Reality (XR) and Machine Learning (ML), in combination, for personalized CIPN sensorimotor rehabilitation. We expect that PROMISE will reduce the severity of the weakened or absent reflexes or the loss of balance control through personalized quantification of deficits and optimal compensation. All these with an affordable, adaptive, and accessible platform for both clinical and home-based rehabilitation.
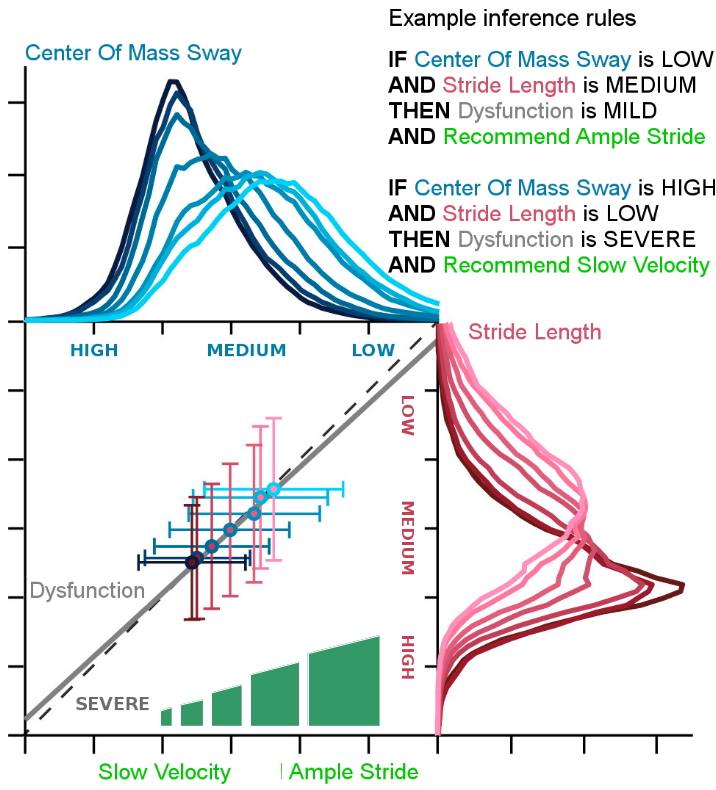
The core element is the inference system capable of translating the assessment into actionable insights and recommendations for the patient in both clinical and home-based rehabilitation deployment of PROMISE.
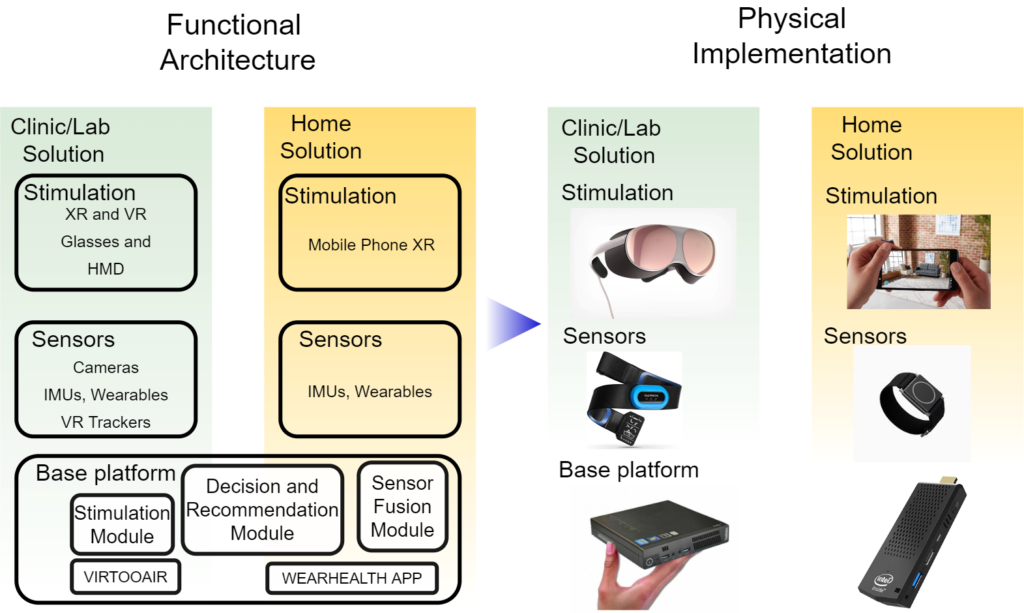
As a technical innovation, PROMISE will offer a platform which can be used in both clinical (laboratory) and home rehabilitation. PROMISE will use a combination of affordable wearables (i.e. IMU, EMG, HR) and recommend the best configuration of number and placement of such sensors that capture patient motion peculiarities.
For instance, the laboratory version will comprise the base platform and additionally all sensors and physician dashboards (i.e. cameras, IMUs, wearables, VR trackers) for a high-accuracy assessment and stimulation. The simulation plays an important role in the clinical version as the physician/therapist can guide the patient. The home-based rehabilitation version of the solution will use a limited set of sensors (i.e. IMUs) and focus continuous monitoring with limited / no stimulation. Such a modular design will allow PROMISE to cover the whole rehabilitation spectrum allowing for continuous patient interaction.