PRECISION: Profile Extraction from Clinical Insights for Smart Individualized Oncology
Over the past decades, early diagnosis, new drugs and more personalised treatment have led to impressive increases in survival rates of cancer patients. Yet, chemotherapy-induced peripheral neuropathy (CIPN), one of the most disabling side effects of commonly used chemotherapeutic drugs, is a severe problem in oncology leading to dose reduction, treatment delay or discontinuation. CIPN causes long-lasting disturbances of daily functioning and quality of life in a considerable proportion of patients. With an increasing number of cancer survivors, more attention is being paid to persistent sequelae of tumour treatment and supportive measures used as adjuncts to mainstream cancer care to control symptoms and enhance well-being.
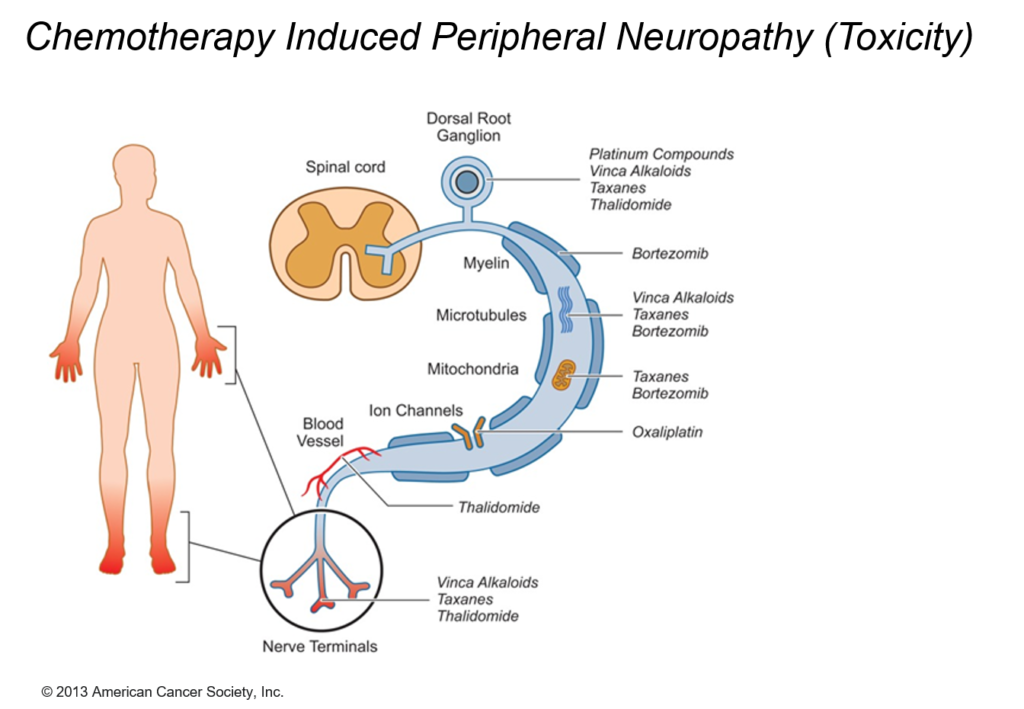
CIPN describes the damage to the peripheral nervous system incurred by a patient who has received a chemotherapeutic agent that is known to be neurotoxic. Independent of the mechanisms of action, the targeted impact of such agents is on axonal transmission with consequences leading up to neuronal apoptosis.
Computational Oncology Approach
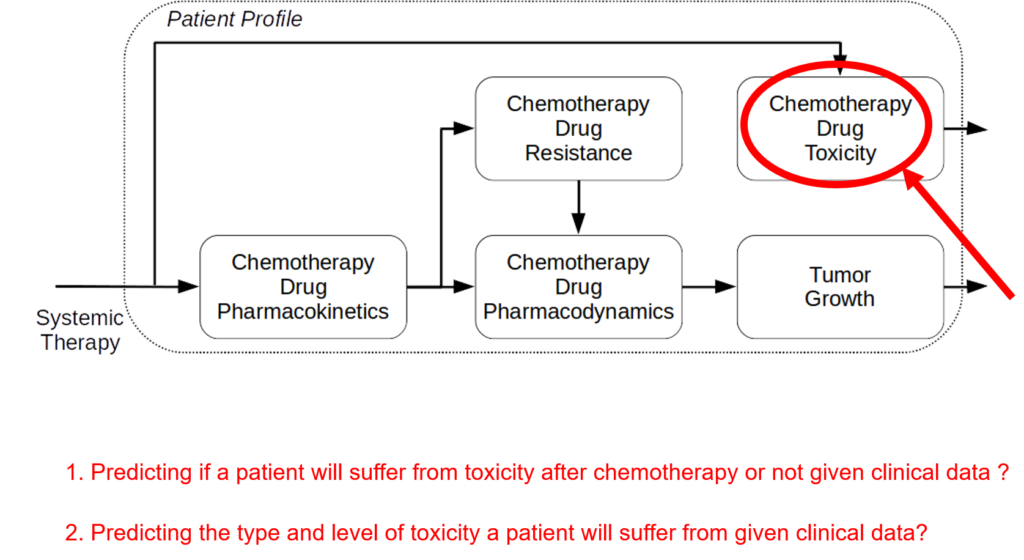
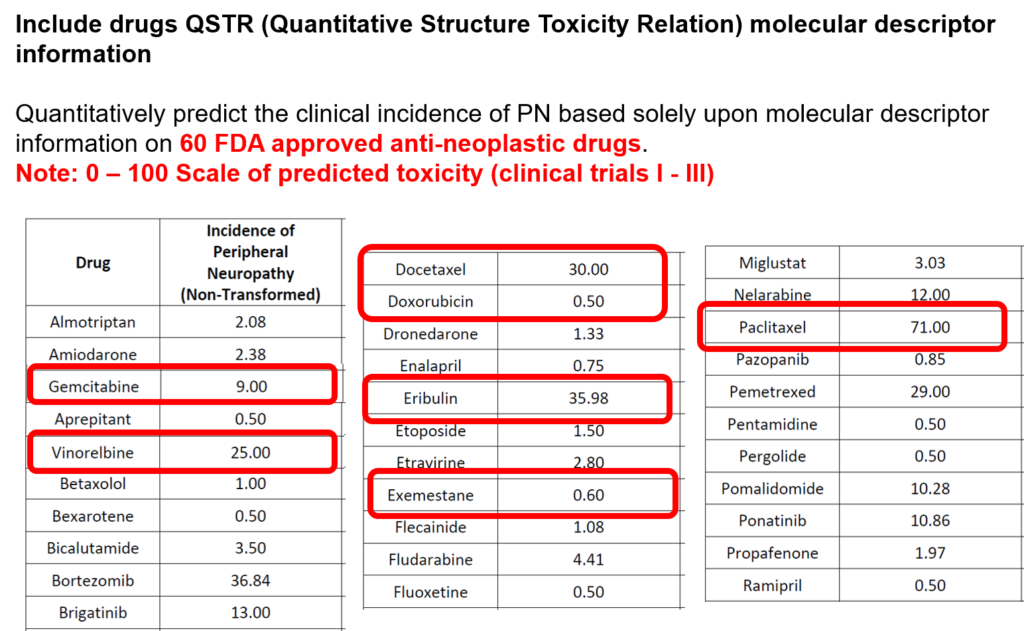
The incidence of peripheral neuropathy differs significantly across chemotherapeutic agents. The prevalence and severity of CIPN are dependent upon several factors relating to both drug pharmacokinetics (e.g., PK, cumulative dose and treatment duration) and pharmacodynamics (e.g., PD, mechanisms of toxicity and patient characteristics). Typically, in vitro and in vivo tests are performed to investigate drug toxicity. In comparison to such experimental approaches, computational methods, including machine learning and structural alerts, have shown great advantages since they are green, fast, cheap, accurate, and most importantly they could be done before the patient receive the therapy. Although such approaches exploit patient-specific risk factors that have been associated with the susceptibility for developing CIPN.
We are aware of no publicly available, validated in silico system to predict the development of CIPN in individual patients. The goal of this project is to develop a clinical decision-support system to predict CIPN development. Using simulation, mechanistic modelling, and machine learning we will develop and validate predictive models to quantify the risk of developing CIPN and deploy a standalone clinical decision-making system that will improve cancer treatment and survivorship care planning.
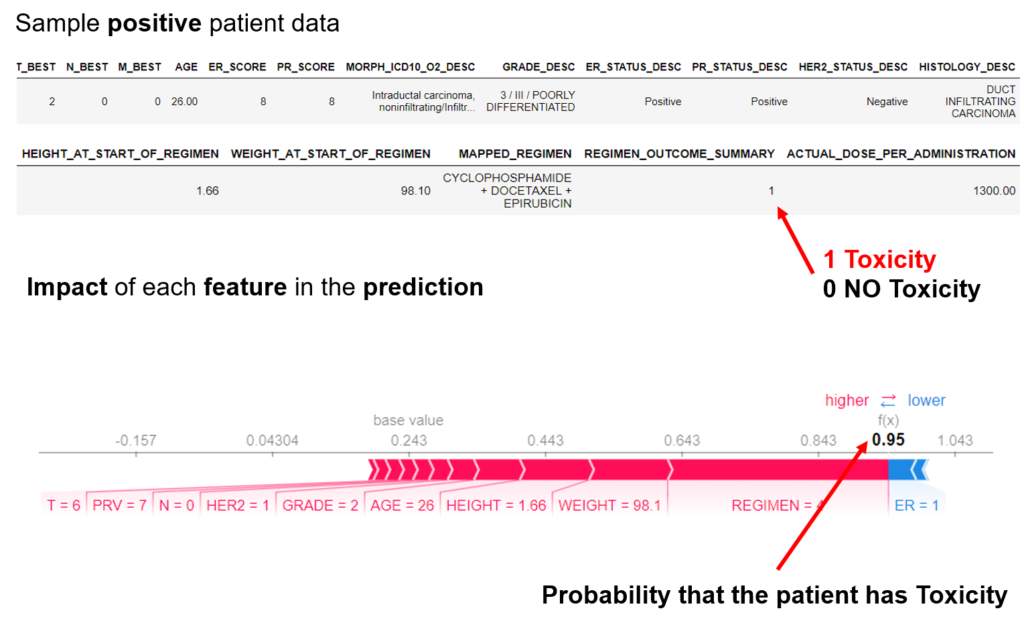
The project proposes a new digital intervention to support clinical oncologists in designing CIPN-aware cancer therapies. Understanding low-level tumor biology and patient genetics can provide unique insights that were previously impossible, to identify genetic predictors of CIPN. Correlating such information with models of tumor growth and patient’s clinical peculiarities, the system can isolate the most relevant cancer response patterns to adjust therapy parameters. The overall goal of the project is to fuse the insights gained at each of these levels in order to build a decision support system that predicts the development of CIPN in a particular patient. The system will learn the governing drug PK/PD differential equations directly from patient data by combining key pharmacological principles with neural ordinary differential equations refined through simulations for improved temporal prediction metrics. Furthermore, by incorporating key PK/PD concepts into its architecture, the system can generalize and enable the simulations of patient responses to untested dosing regimens. These emphasize the potential such a system holds for automated predictive analytics of patient drug response. The end result of the project will be a simple app that will collect physician inputted patient clinical parameters and eventually health records and will provide, given the therapy drug choice a prediction of the severity of CIPN before the chemotherapy regimen. Additionally, the physician and patient will get interpretable insights and a clear explanation of how each of the factors affects the prognosis. The entire processing and data aggregation and learning will happen in the cloud using GDPR-aware data handling.
The project is based on large-scale data fusion that provides a multi-faceted profile of the patient, drugs, tumor and how they interact. This will finally support personalized intervention and adaptive therapies that maximise efficacy and minimize toxicity.